Link to my master's thesis PDF: Fast Surface Reconstruction and Mesh Correspondence for Longitudinal 3D Brain MRI Analysis (20 MB)
In my master's thesis I worked on improving existing cortical surface reconstruction methods. Specifically, my work focused on more consistent mesh representations, particularly vertex locations, across time points for the same patient. This approach allows for more accurate vertex-wise longitudinal analysis.
But what are the brain's cortical surfaces?
Cortical Surfaces
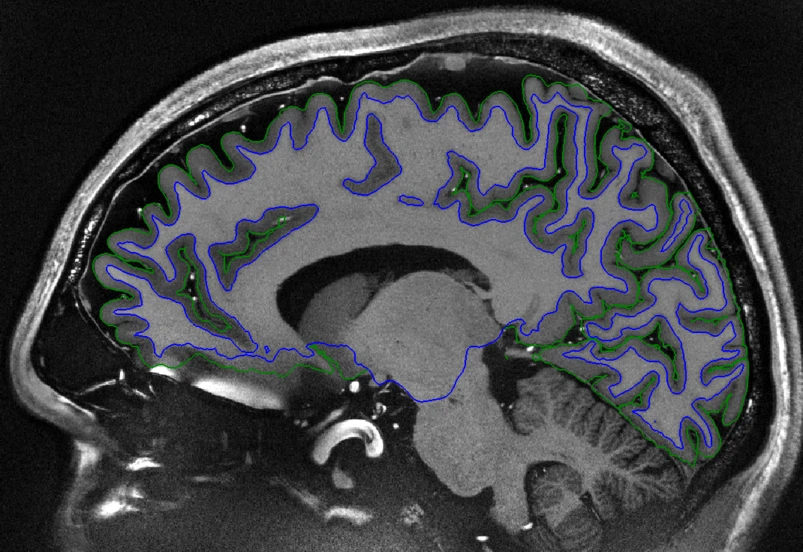
Sagittal view of a T1-weighted human brain MRI scan. Creative Commons Attribution 4.0 International License - Source
The cerebral cortex is the outer part of the human brain and responsible for many important cognitive functions. It consists of a thin layer of gray matter covering the outermost surface of the brain with a thicker layer of white matter underneath.
Declines in cortical thickness and volume are associated with many neurological diseases such as Alzheimer's disease. Because of this, measuring cortical thickness and volume is an important task for studying and detecting neurological diseases.
To measure the thickness, two boundaries of the cortex are defined: the white matter surface (blue in the above image) and the pial surface (green in the above image). Interestingly, these surfaces have a spherical topology, meaning that if you could stretch them out, you could form a ball out of them.
These surfaces are typically represented as triangular meshes. Triangular meshes consist of vertices in 3D space, and edges and faces connecting these vertices. Besides allowing you to 3D print your brain, this mesh representation is useful for precise measurements of cortical thickness and volume.
Note: All meshes shown here were reconstructed from scans from here published under a Creative Commons Attribution 4.0 International License.
Cortical Surface Reconstruction
The conversion of a grayscale voxel-based (3D equivalent of pixels) MRI scan to a mesh representation is called cortical surface reconstruction.
The standard tool for cortical surface reconstruction is FreeSurfer. It's a dusty old tool written in C++ whose main interface consists of a 10.000 line tcsh script 😥. However, due to its popularity, it's well-tested and validated.
Reconstructing a single patient with FreeSurfer may take multiple hours, making reconstruction in large-scale studies difficult.
As with many fields, deep learning has made its way into cortical surface reconstruction. Deep learning-based methods are typically much faster than FreeSurfer and predict the meshes directly from the MRI scan within seconds. Of course, the methods are not perfect yet. Some problems are (the extent of which depends on the method):
- irregular/self-intersecting faces
- white matter/pial surface intersections
- no real ground truth data (typically FreeSurfer meshes are used as silver standard)
- lack of reconstruction accuracy
- topological errors (holes, "bridges", etc.)
- staircase artifacts
- consistent over-/underestimation of cortical thickness
The method that my thesis is based on is called V2C-Flow, a deep surface reconstruction method developed by the Artificial Intelligence in Medical Imaging Lab at TU Munich. The corresponding paper is currently in review (as of December 2023). V2C-Flow deforms a set of template meshes to the target pial/white matter surfaces. This is achieved by a U-Net that segments the MRI scan into white matter, gray matter and background and a set of graph convolution blocks in combination with neural ODEs that predict the displacement of each of the template vertices. The U-Net latent activations are used to guide the deformation process. V2C-Flow is state-of-the-art in terms of reconstruction accuracy, but suffers from points 1, 3, and, 7 above.
Longitudinal Mesh Alignment
In many studies, multiple scans are acquired from the same patient over longer periods of time, e.g. to study the progression of a disease. These studies are called longitudinal studies.
For vertex or region-wise tracking of changes over time, it is crucial that the meshes of the same patient at different time points are aligned well, meaning that the same vertex in the first mesh corresponds to the same vertex in the second mesh and that both vertices are located at the same anatomical position. This type of alignment also allows you to smoothly interpolate between the meshes, which is useful for visualizing the progression of a disease.
As a first requirement, the meshes need to have the same number of vertices and the same connectivity. Otherwise, it is not possible to identify the same vertex in both meshes. This is by default not the case for FreeSurfer, but is the case for V2C-Flow, due to the shared template mesh.
However, FreeSurfer ships with a dedicated longitudinal pipeline that both creates meshes with the same number of vertices and aligns them. V2C-Flow meshes, on the other hand, are only roughly aligned. The goal of my thesis was to find a new method for producing well-aligned meshes in a deep learning-based manner.
Approach
During my thesis, I tried many different approaches to align longitudinal meshes. Some naive ideas like L2-losses or nearest-neighbor resampling did not work well, but after tinkering with some more approaches, the final method, which is called V2C-Long, was decided on. It is based on the following idea:
- Use a standard V2C-Flow model to reconstruct all meshes of the same patient at different time points.
- Create patient-wise median meshes for both pial and white matter surfaces by aggregating the vertices of the respective meshes from all time points of the same patient.
- Use a second V2C-Flow model to deform the median mesh to each of the individual meshes.
This approach requires double the inference time of V2C-Flow, but is still much faster than FreeSurfer (by a factor of over 1000x). The median template has two advantages: First, it removes outliers, which reduces the self-intersections in the patient-wise meshes and in the final meshes. Second, the median template is very close to each of the final meshes (when compared to the standard, patient-agnostic template), leading to very small GCN deformations of the template vertices. As a consequence, the predicted vertices are all located at similar positions.
Result
Not only does my method significantly improve the alignment of the meshes, it also reduces the number of self-intersections and improves the reconstruction accuracy! This is like a triple win! 🎉
Below you can find some example meshes, rendered in a way that shows the actual mesh structure. For all methods, a small part of the white matter surface is shown for two reconstructions of the same patient. Note that the FreeSurfer mesh covers a larger area of the surface. The same set of faces (i.e. the corresponding faces/edges/vertices) were selected.
The V2C-Flow method has a big misalignment/offset between the meshes. This is especially clear at the boundaries. On the other hand, both FreeSurfer and my method (V2C-Long) produce very aligned meshes. In my benchmarks, my method even outperforms FreeSurfer in most cases! The method is still not perfect (this can be seen at some corners that don't match perfectly), but a big improvement when compared to V2C-Flow.

Personal Summary
I really enjoyed working on this project and dipping my toes into a fairly new field for me (medical imaging and mesh processing). Also, the project was more interdisciplinary than stuff I did previously, which was a nice change. I'm also curious if the research will actually help progress the field of cortical surface reconstruction (even if just for a little bit), but this will only be clear in a few years.
I won't return to this field in the near future, but, who knows, maybe some day I'll work on similar topics again.